Research in the Ni Lab
This page lists some of the past projects and on-going future projects. Please feel free to contact taoni@hku.hk for discussion.
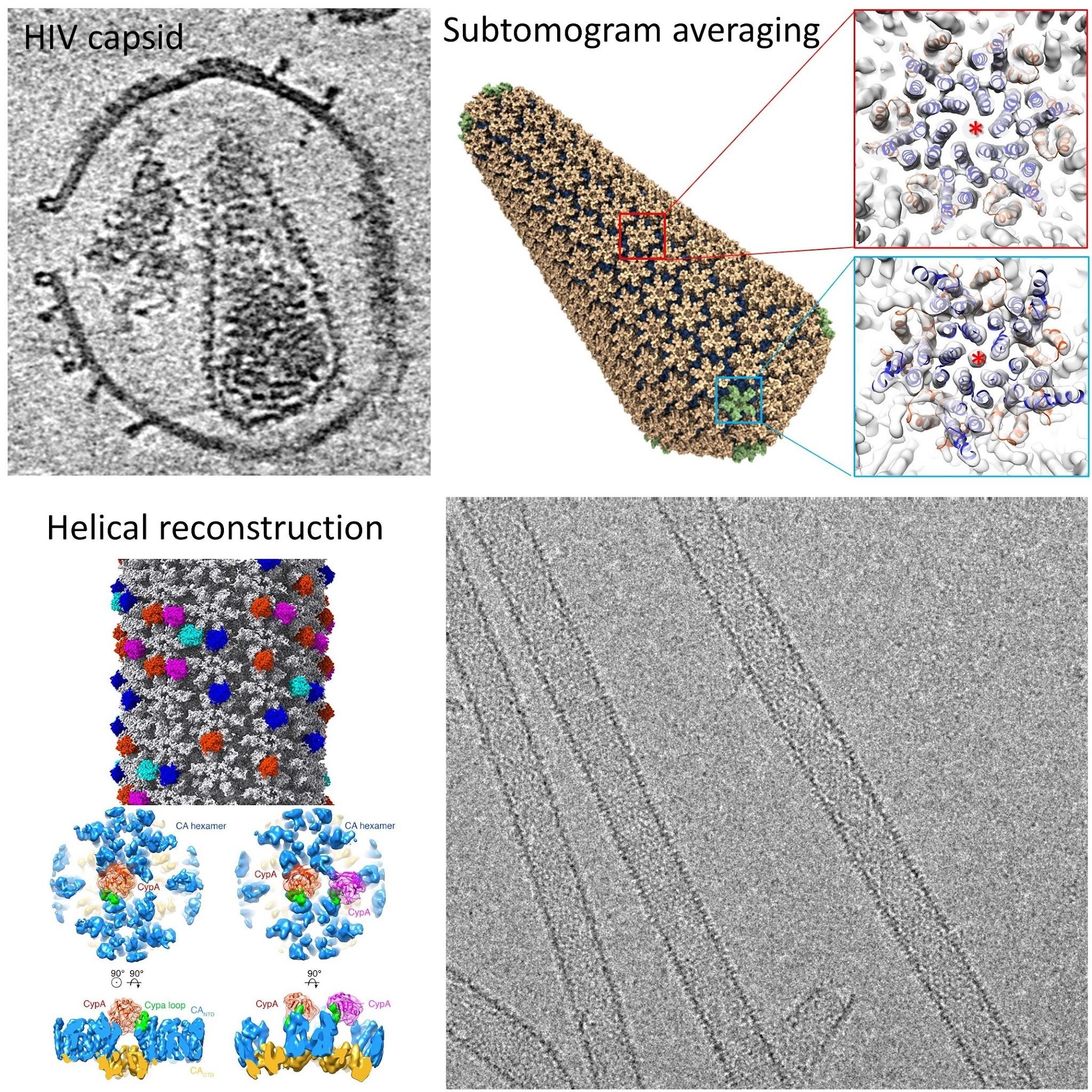
Understanding HIV capsid
HIV capsid plays key roles in HIV replication and is a major platform engaging host dependency and restriction factors. Our previous work determined high-resolution structures of in vitor assembled HIV capsid tube in complex with host cell factor Cyclophilin A, elucidating the mechanism of capsid assembly and curvature sensing (Ni et al 2020 NSMB; Ni et al 2021 Science Advances).
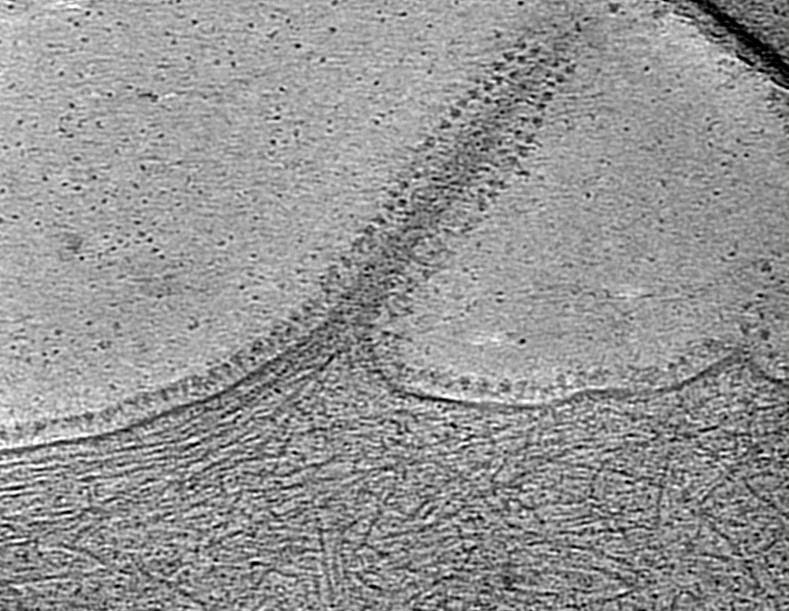
Understanding host-coronavirus interplay using cryoET
Over the last two decades, there were three outbreaks for severe coronavirus infection in humans including the current pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for the Coronavirus Disease 2019 (COVID-19). With the global health and economic burden due to Covid-19, it is increasingly urgent to understand the coronavirus life cycle and pathogenesis in molecular level.
Coronavirus undergoes a complicated life cycle in the host cell and makes a significant rearrangement of host membranes. It transforms host membranes into double-membrane vesicles (DMVs) which may form a framework for viral genome replication by localizing and concentrating the necessary factors and possibly providing protection from host cell defenses. Specifically, the membrane remodelling and formation of DMVs are induced by a few non-structural proteins (Nsp3, Nsp4 and Nsp6), conserved in coronavirus family. These proteins form a putative pore complex on the DMVs, potentially mediating genome transport after new genomes are synthesized and messenger RNAs are produced inside the DMVs. The pore may serve as a scaffold for the assembly of the membrane-associated replication/transcription complex (RTC), a large multi-subunit assembly that is comprised of more than a dozen proteins. We aim to understand the role of Nsp3 pore complex and DMV in coronavirus life cycle.
The project will use a combination of biochemical and structural biology approach. We are looking for candidates who are interested in virology and/or structural biology including cryoEM single particle analysis and cryoET subtomogram averaging.
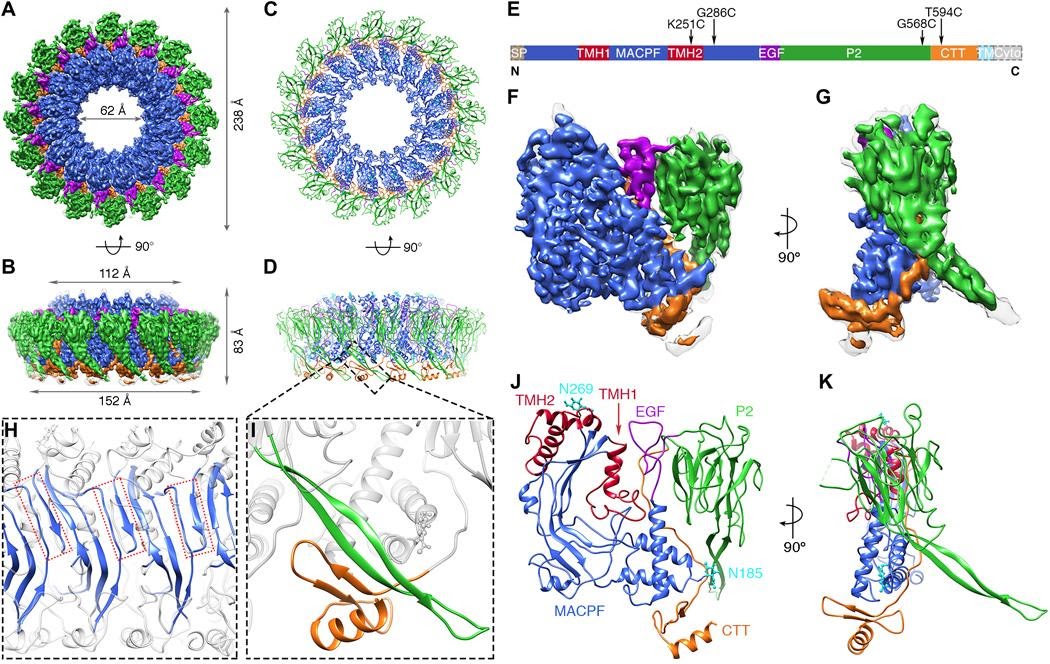
MACPF pore-forming proteins
Membrane Attack Complex (MAC)/perforin family proteins represent an important ans conserved pore-forming proteins involved in immunity and defense. They form giant pores on membrane, following a dramatic transition between prepore to pore. We have determined several key structures by X-ray crystallography and cryoEM, cryoET subtomogram averaging (Ni et al 2018, Science Advances, Ni et al 2020, Science Advances).
We are continuing to solve the puzzle of pore-forming proteins, focusing on perforin-2. More interesting story is comming soon…
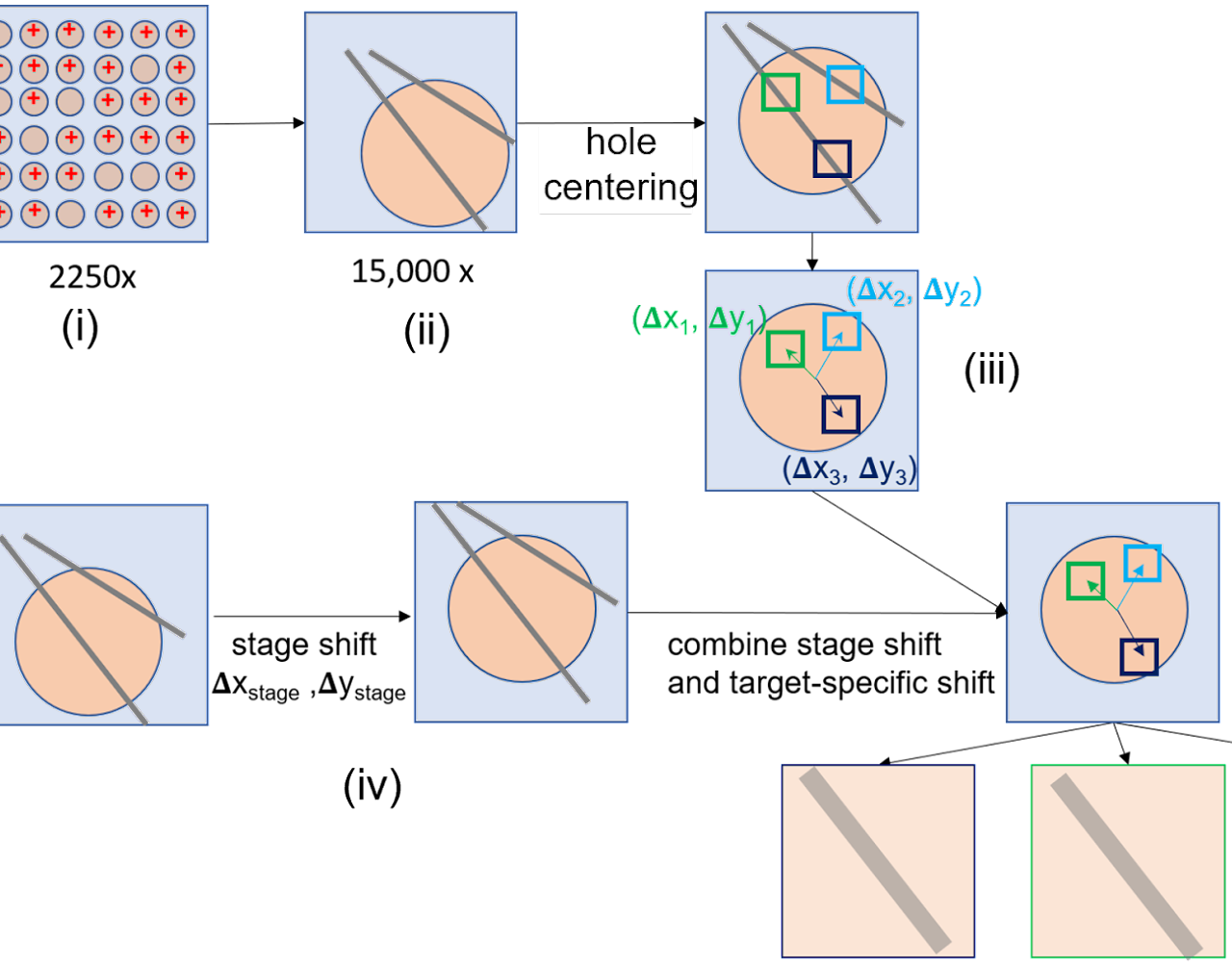
Method development in cryoET
Cryo-electron tomography (cryoET) has gained increasing importance to study molecular architectures of viruses, bacteria and cellular components in situ. It can provide structures of macromolecular complexes in situ and in cellular context at or below subnanometer resolution. Combined with cryo-focused ion beam (cryo-FIB) milling for sample preparation this has led to unprecedented insights into the inner working of molecular machines in their native environment, as well as their functional relevant conformations and spatial distribution within biological cells or tissues. Despite its rapid development in recent years, many hardware and software are still evolving to address the increasing challenges due to the complexity of biological systems.
We are looking for candidates with strong interest in data processing and software development. The specific projects will be determined together with PI and student and potential directions include but not limit to:
- Development of hardware and software for automated high-throughput cryoET data collection and pre-processing;
- Method development for cryoET data analysis and subtomogram averaging, such as using Artificial Intelligence (AI) based approach for target recognition;
- Methods development and optimization for cryogenic correlative light and electron microscopy (cryo-CLEM).